The skin is the largest organ and forms the outermost layer of the human body. There-fore, the skin is more easily exposed to toxic environmental agents, particularly ultraviolet (UV) radiation, than are other tissues, except for the eyes. Human skin consists of the epidermis, dermis and hypodermis. The most abundant cells in the dermis layer are human dermal fibroblasts (HDFs), which contribute to skin firmness and elasticity by upregulating collagen synthesis. However, chronic and continuous exposure to UV radiation leads to upregulation of matrix metalloproteinases (MMPs), collagen degradation and wrinkle formation, which are the main features of skin aging. In addition, UV radiation generates reactive oxygen species and DNA damage that induce cellular senescence and apoptosis in HDFs. Furthermore, exposure to UV radiation is an important causative factor for skin diseases, such as photodermatoses, actinic keratosis and skin cancer. However, UV-in-duced skin aging is not an inevitable natural consequence, but is due to extracellular factors; in other words, the aging process is controllable by avoidance of UV irradiation.
Accumulating evidence indicates that the application of a chemical sunscreen, a commonly known as sun creams with an adequate sun protection factor (SPF), is able to decrease UV-induced DNA damage, such as the formation of pyrimidine dimers, in hu-man skin. However, the eff ectiveness of a sunscreen has been challenged by new research. Notably, sunscreen is only eff ective in preventing UV-induced DNA damage when used regularly. Furthermore, a recent study demonstrated that sunscreens with superior UVA protection and UVB SPF 50 delayed the onset of UV radiation-in-duced melanoma skin cancer, but provided only partial protection, indicating that UV light can sneak past the sunscreen and cause long-term DNA damage, even in case of an SPF 50 sunscreen. These results indicate that sunscreen-mediated UV blockage is not 100 % protective against UV radiation, and has no lasting UV-protective effect. Sunscreen exerts only a physical protective effect within its given SPF time, not a biological protective effect against UV radiation in human skin.
Thus, identifi cation of novel reagents exerting a biological UV protective effect in skin cells is an important future strategy for preventing UV-induced skin aging. To this end, we have developed a novel large-scale screening system, the DNA repair regulating material discovery (DREAM) system. This system facilitates the high-throughput identifi cation of materials that effectively protect against and/or repair UV-induced DNA damage in HDFs
EXPERIMENTAL
Cells and reagents
Recombinant lentivirus construction
UV irradiation
Cytotoxicity assay
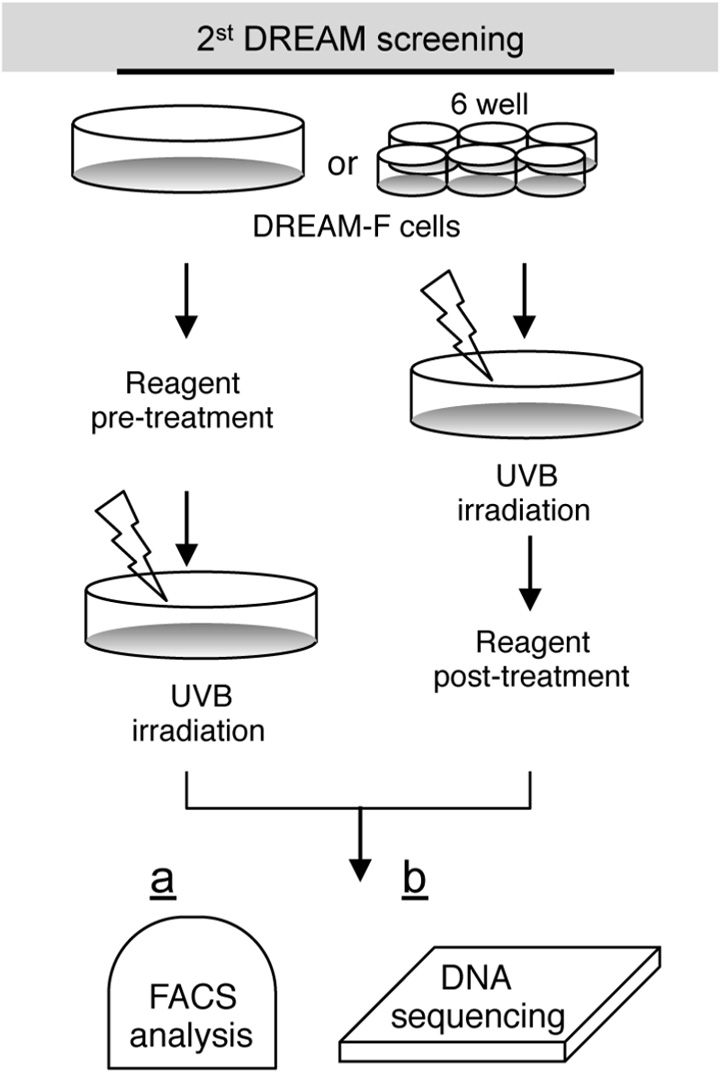
Flow cytometric analysis
Luciferase assay
Statistical analysis
RESULTS AND DISCUSSION
Construction of lentiviruses containing the luciferase and HPRT genes
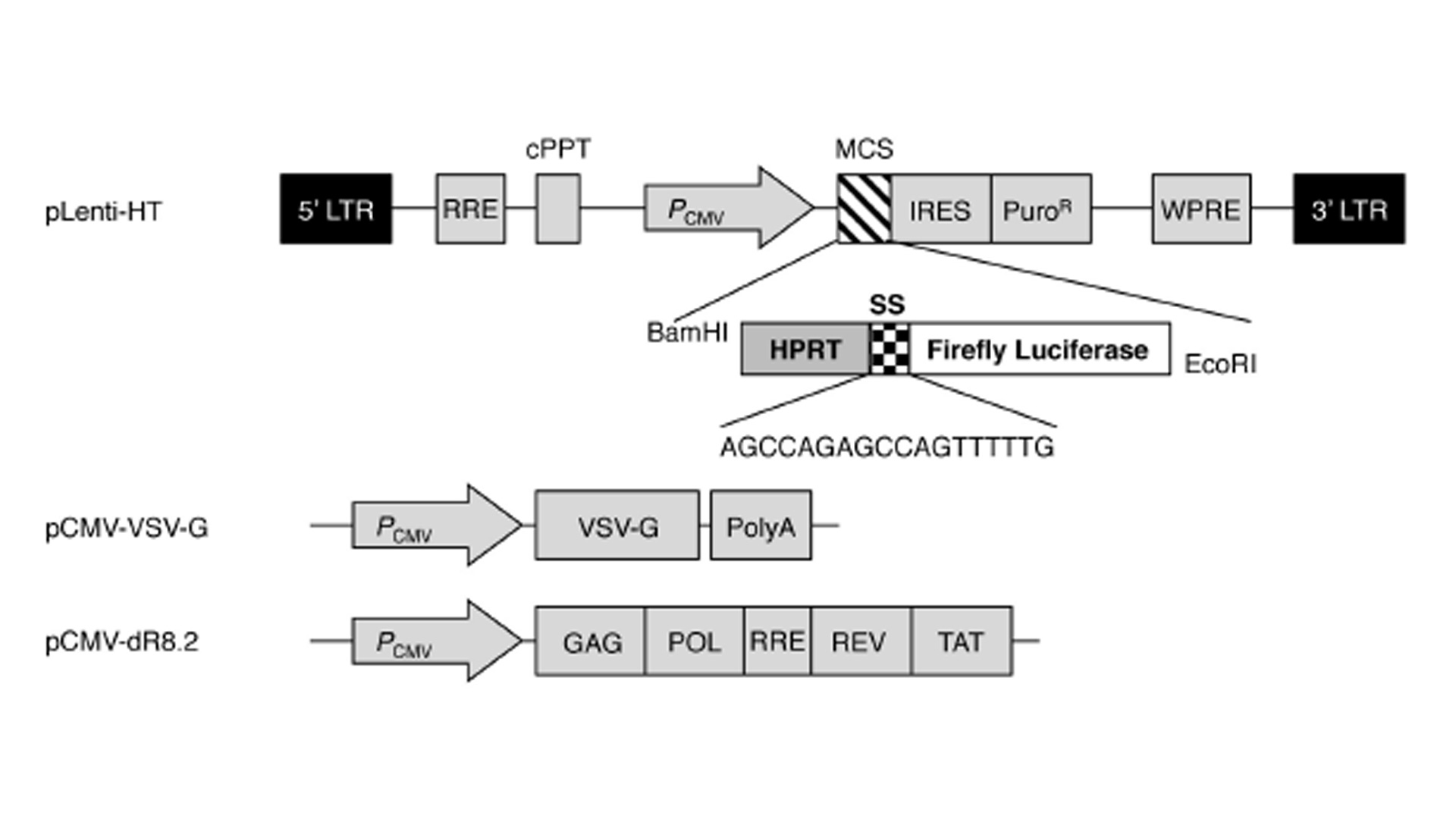
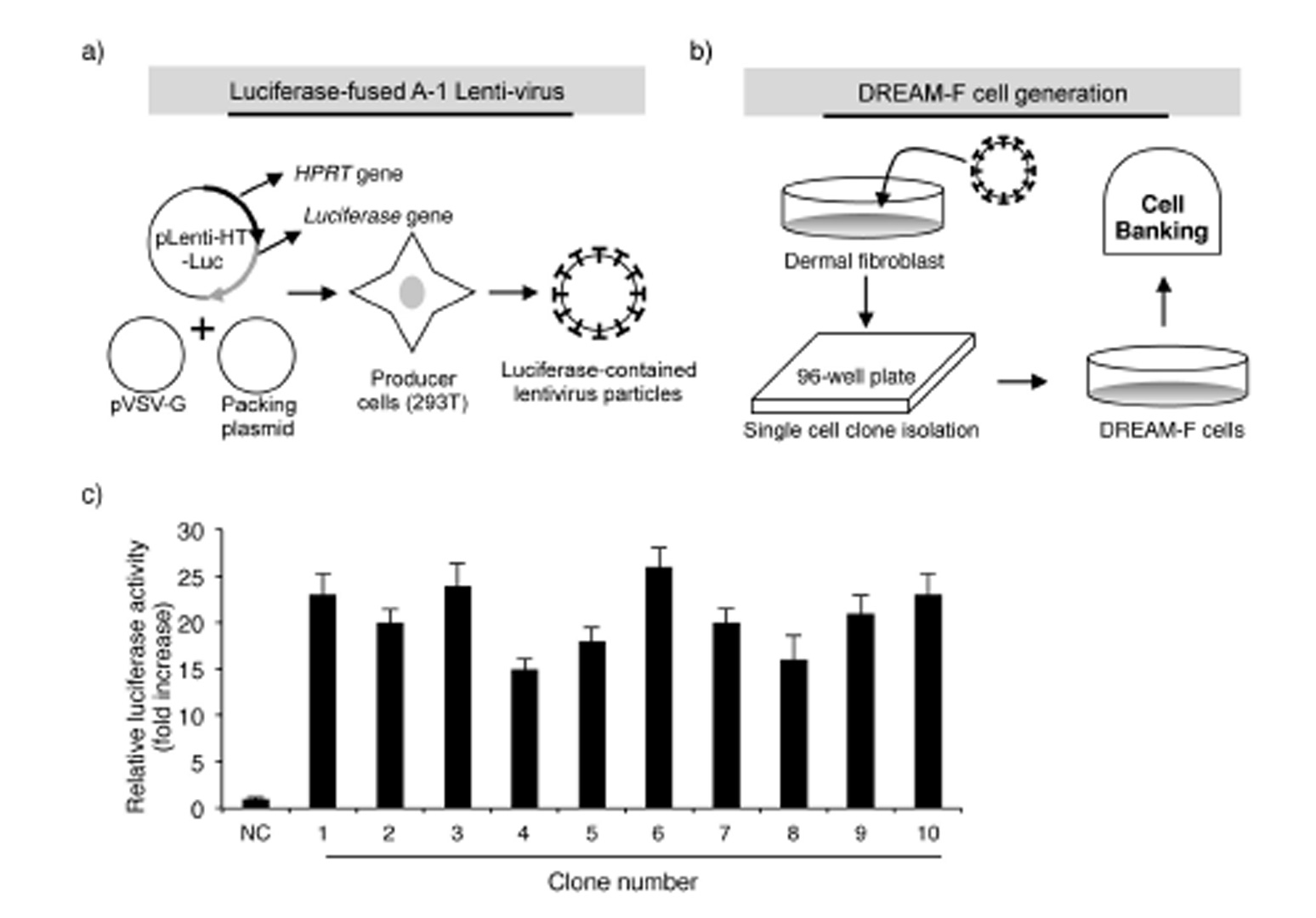
Based on the above results, we aimed to develop a UV-induced DNA damage sensor system using human dermal fibroblasts. First, the UV-induced reduction in transcription and altered mutant protein level were evaluated using a luciferase system. The luciferase assay is a technique with a diverse range of applications in molecular biology and we confirmed that UV irradiation significantly reduced the activity of luciferase in luciferase vector-transfected NHDFs (data not shown). Next, to render the luciferase system more vulnerable against UV radiation, we inserted sensor sequences (SSs; 5’-AGCCAGAGC-CAGTTTTTG-3’) upstream of the luciferase ORF sequence. A study by Kreimer-Erlacher et al. Showed that in CC-containing sequences, the latter C was more commonly substituted with T than the former C (CC -> CT transition). Therefore, following UV irradiation, a stop codon is likely to be created in the ‘AGCCAG’ sequence in the SS (AGCCAG -> AGCTAG). The sequence of TTTTT in the SS has the potential to develop CPDs following UV irradiation. Furthermore, to analyze the exact number of mutations following UV irradiation, the housekeeping gene HPRT was cloned into the N-terminal region of the SS. A study of Kappes at al. Showed that irradiation with 100, 200 and 400 J m-2 UVB resulted in the generation of roughly 10, 23 and 59 mutations in the HPRT gene in human fibroblasts. Therefore, in summary, the HPRT-SS-luciferase fusion sequence was used to evaluate the level of UV-induced damage. To generate NHDFs expressing the HPRT-SS-luciferase protein, we used the lentiviral system, which stably infects dividing and non-dividing human cells. As shown in Fig. 1, the HPRT-SS-luciferase sequence was inserted between the CMV promoter and the IRES sequence in the pLenti-HT vector. The ORF region of firefl y luciferase was cloned into the EcoRI site of pLenti-HT. pCMV-VSV-G is a VSV-G coding plasmid, which allowed the production of viral particles. pCMV-dR8.2 is a packaging plasmid, which encodes 4 genes, including Gag, Pol, Rev, and Tat. Subsequently, lentivirus particles were generated by transfecting 293T cells with the recombinant pLenti-HT, packaging vector pCMV-dR8.2 and pCMV-VSV-G. Aft er 48 h of transfection, recombinant lentivirus particles were collected by filtration (Figs. 2a,b).
Fig. 1
Vector maps of pLenti-HT-luciferase, the packaging plasmid and the envelope plasmid. LTR-long terminal repeat sequence, RRE, rev response element; cPPT, central polypurine tract; PCMV, cmv promoter; MCS, multiple cloning sites; IRES, internal ribosome entry site; PuroR, puromycin resistant gene; WPRE, woodchuck hepatitis virus post-transcription regulatory element; VSV-G, vesicular stomatitis virus GP; PolyA, poly-A sequence.
Generation of HPRT-SS-luciferase-expressing DREAM-F cells
Fig. 2
Schematic diagram of luciferase-expressing lentivirus production and DREAM-F cell generation: a) Luciferase-expressing lentiviruses were produced by transfecting 293T cells with the pLenti-HT-HPRT-SS-luciferase, pCMV-VSV-G and pCMV-dR8.2 plasmids; b) Establishment of luciferase-fused WS1 cells (DREAM-F cells); c) Identification of a cell clone with the highest expression level of luciferase. The results are expressed as the means ± SD from three separate experiments; NC – nega-tive control. Significant difference compared with control WS1 cells (*p < 0.05).
Establishing a system to identify novel reagents regulating UVB-induced DNA damage using DREAM-F cells
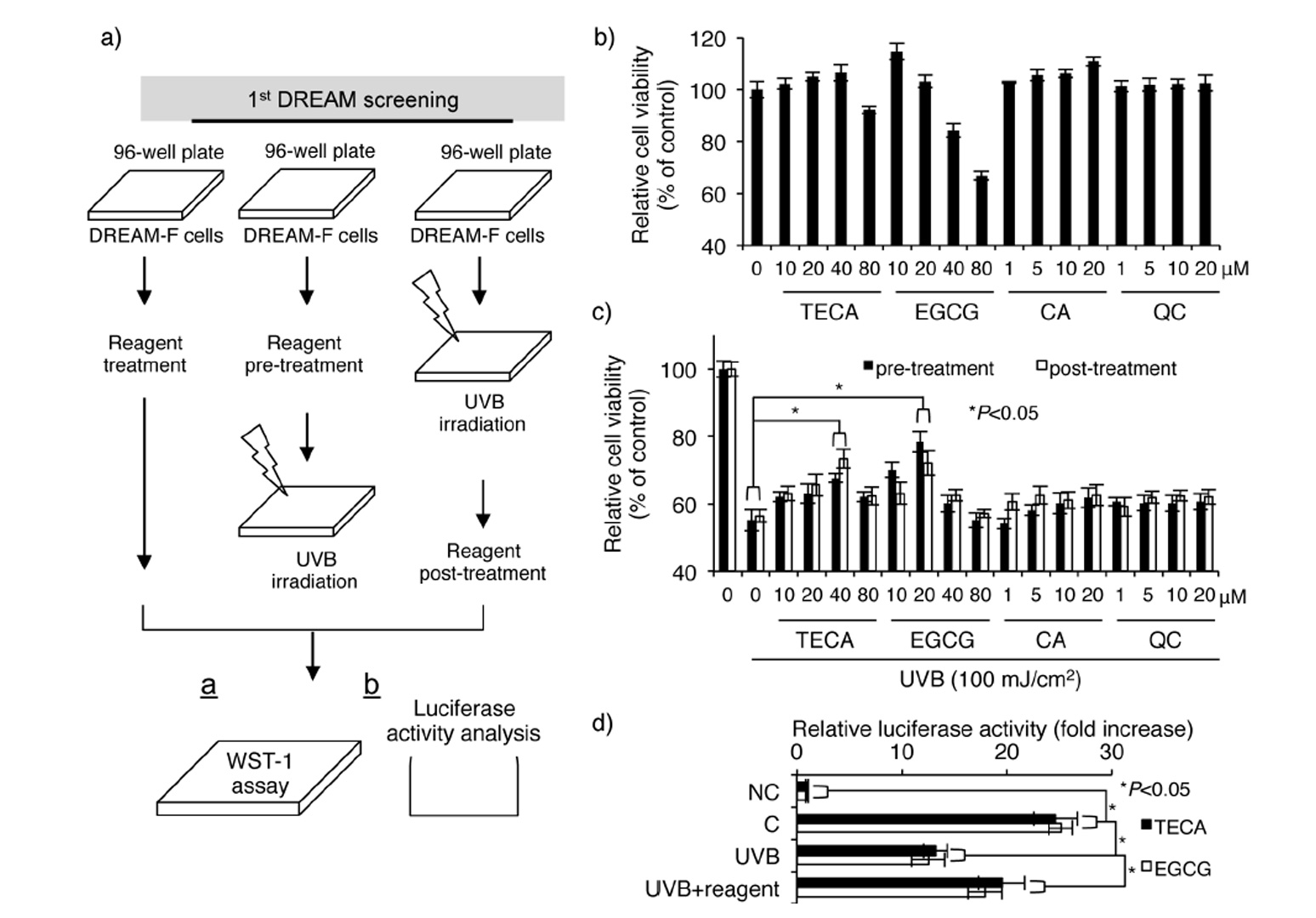
To validate this system, we used four types of phytochemicals: titrated extract of Centella asiatica (TECA), epigallocatechin gallate (EGCG), caffeic acid (CA), and quercetin (QC). Our group recently found that TECA exerts a UV-protective effect in human dermal fi bro-blasts by downregulating UV-induced cellular and DNA damage (14). Furthermore, it was demonstrated that EGCG has a UV-protective effect in human dermal fibroblasts (15). CA and QC are known to exert inhibitory effects against UV-induced oxidative stress (16, 17). As shown in Fig. 3b, relatively high concentrations of TECA and EGCG exhibited cytotoxicity in DREAM-F cells, whereas CA and QC showed no cytotoxicity. Interestingly, UV-protective and UV-induced cellular damage repair effects were only present in TECA- and EGCG-treated DREAM-F cells (Fig. 3c). Further experiments showed that TECA and EGCG inhibited UV-induced loss of luciferase activity in DREAM-F cells (Fig. 3d), indicating that these two reagents have a UV-protective and UV-induced DNA damage repair effects in human dermal fibroblasts. Our previous study demonstrated that TECA inhibits UVB-induced cellular toxicity; however, we also found that TECA directly inhibited UVB-in-duced DNA damage using our DREAM system. Other reports showed that EGCG reduced UVB-induced oxidative DNA damage in living skin equivalents (18). Although previous reports demonstrated CA- and QC-mediated anti-oxidative effects in hairless mice and lymphocytes, these effects might be cell-specifi c or may not be directly involved in the loss of cell viability mediated by UV radiation (16, 17). Overall, our results indicate that the DREAM system is useful for the identification of novel reagents regulating UV-induced DNA damage in human dermal fibroblasts.
Other potential applications of the DREAM system
Fig 3
Schematic diagram and confirmation of the first DREAM screening system: a) Strategy for the identification of putative reagents showing protective effect against UVB radiation and regulatory effect on UVB-induced loss of luciferase activity; b) Evaluation of cytotoxicities for TECA, EGCG, CA and QC; c) Evaluation of the protective effect against UVB-induced loss of cell viability. Significant difference compared to UVB-irradiated DREAM-F cells (*p < 0.05); d) Evaluation of protective effect against UVB-induced downregulation of luciferase activity (mean ± SD, n = 3). Significant difference compared to control WS1 cells and UVB-irradiated DREAM-F cells (*p < 0.05).
CONCLUSIONS
Our aim was to develop a large-scale and high-throughput screening system for identifying novel reagents that protect against or repair UV-induced DNA damage in human dermal fibroblasts. For this purpose, we used a 96-well-based screening system using DREAM-F cells. In conclusion, our study demonstrates that the hybrid luciferase gene can be used for evaluating the level of DNA damage caused by UV irradiation in human dermal fibroblasts. Furthermore, we generated DREAM-F cells using lentiviruses. This hybrid cell offers a large-scale and high-throughput screening system for novel anti-aging reagent under false-positive-free conditions. Also, we validated this DREAM system using TECA, EGCG, CA, QC reagents, and found that TECA and EGCG are novel phytochemicals showing a direct inhibitory effect on UV-induced DNA damage in dermal fibroblasts. Therefore, we believe that the DREAM system is appropriate for identification of novel pharmaceutical reagents showing anti-photoaging or anti-photocarcinogenesis in the future.
Abbreviations, acronyms and symbols. – DREAM system – DNA repair regulating mate-rial discovery system, HPRT – hypoxanthine phosphoribosyl transferase, HDFs – human dermal fibroblasts, MMPs – matrix metalloproteinase, SPF – sun protection factor, CPDs – cyclobutane pyrimidine dimers, TECA – titrated extract of Centella asiatica, EGCG –epigallocatechin gallate, CA – caff eic acid; QC – quercetin.
Acknowledgements. – Lenti-HT vector was kindly provided by Prof. Jae Ho Lee of Kwandong University, Seoul, Republic of Korea. The paper was supported by the KU Research Professor Pro-gram of Konkuk University and a grant of the Korean Health Technology R&D Project (Grant No. HN13C0080), Ministry of Health & Welfare, Republic of Korea.
REFERENCES
- N. Philips, S. Auler, R. Hugo and S. Gonzalez, Benefi cial regulation of matrix metalloproteinases for skin health, Enzyme Res. 2011, article ID 427285; DOI: 10.4061/2011/427285.
- O. Friedman, Changes associated with the aging face, Facial Plast. Surg. Clin. North. Am. 13 (2005) 371–380; DOI: 10.1016/j.fsc.2005.04.004.
- B. Wang, Photoaging: a review of current concepts of pathogenesis, J. Cutan. Med. Surg. 15 (Suppl. 1) (2011) S374–S377; DOI: 10.2310/7750.2011.00006.
- M. A. Farage, K. W. Miller, P. Elsner and H. I. Maibach, Intrinsic and extrinsic factors in skin age-ing: a review, Int. J. Cosmet. Sci. 30 (2008) 87–95; DOI: 10.1111/j.1468–2494.2007.00415.x.
- A. Fourtanier, D. Moyal and S. Seite, Sunscreens containing the broad-spectrum UVA absorber, Mexoryl (R) SX, prevent the cutaneous detrimental eff ects of UV exposure: a review of clinical study results, Photodermatol. Photoimmunol. Photomed. 24 (2008) 164–174; DOI: 10.1111/j.1600-0781.2008.00365.x.
- S. E. Freeman, R. D. Ley and K. D. Ley, Sunscreen protection against UV-induced pyrimidine dimers in DNA of human skin in situ, Photodermatology 5 (1988) 243–247; DOI: 10.1046/j.0022-202x.2001.01580.x.
- T. J. Phillips, J. Bhawan, M. Yaar, Y. Bello, D. Lopiccolo and J. F. Nash, Eff ect of daily versus inter-mitt ent sunscreen application on solar simulated UV radiation-induced skin response in humans, J. Am. Acad. Dermatol. 43 (2000) 610–618; DOI: 10.1067/mjd.2000.107244.
- M. Al Mahroos, M. Yaar, T. J. Phillips, J. Bhawan and B. A. Gilchrest, Eff ect of sunscreen applica-tion on UV-induced thymine dimers, Arch. Dermatol. 138 (2002) 1480–1485; DOI: 10.1001/arch-derm.138.11.1480.
- A. Viros, B. Sanchez-Laorden, M. Pedersen, S. J. Furney, J. Rae, K. Hogan, S. Ejiama, M. R. Girott i, M. Cook, N. Dhomen and R. Marais, Ultraviolet radiation accelerates BRAF-driven melanoma-genesis by targeting TP53, Nature 511 (2014) 478–482; DOI: 10.1038/nature13298.
- C. P. Selby, R. Drapkin, D. Reinberg and A. Sancar, RNA polymerase II stalled at a thymine dimer: footprint and eff ect on excision repair, Nucleic Acids Res. 25 (1997) 787–793.
- H. Soehnge, A. Ouhtit and O. N. Ananthaswamy, Mechanisms of induction of skin cancer by UV radiation, Front Biosci. 2 (1997) 538–551.
- U. P. Kappes, D. Luo, M. Pott er, K. Schulmeister and T. M. Runger, Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells, J. Invest. Dermatol. 126 (2006) 667–675; DOI: 10.1038/sj.jid.5700093.
- H. Kreimer-Erlacher, H. Seidl, B. Back, L. Cerroni, H. Kerl and P. Wolf, High frequency of ultraviolet mutations at the INK4a-ARF locus in squamous cell carcinomas from psoralen-plus-ultraviolet-A-treated psoriasis patients, J. Invest. Dermatol. 120 (2003) 676–682; DOI: 10.1046/j.1523-1747.2003.12085.x.
- I. S. An, S. An, S. M. Kang, T. B. Choe, S. N. Lee, H. H. Jang and S. Bae, Titrated extract of Centella asiatica provides a UVB protective eff ect by altering microRNA expression profi les in human dermal fi broblasts, Int. J. Mol. Med. 30 (2012) 1194–1202; DOI: 10.3892/ij mm.2012.1117.
- I. S. An, S. An, S. Park, S. N. Lee and S. Bae, Involvement of microRNAs in epigallocatechin gallate-mediated UVB protection in human dermal fi broblasts, Oncol. Rep. 29 (2013) 253–259; DOI: 10.3892/or.2012.2083.
- R. Casagrande, S. R. Georgett i, W. A. Verri, Jr., D. J. Dorta, A. C. dos Santos and M. J. Fonseca, Protective eff ect of topical formulations containing quercetin against UVB-induced oxidative stress in hairless mice, J. Photochem. Photobiol. B 84 (2006) 21–27; DOI: 10.1016/j.jphotobiol.2006.01.006.
- N. R. Prasad, K. Jeyanthimala and S. Ramachandran, Caff eic acid modulates ultraviolet radiation-B induced oxidative damage in human blood lymphocytes, J. Photochem. Photobiol. B 95 (2009) 196–203; DOI: 10.1016/j.jphotobiol.2009.03.007.
- S. Y. Kim, D. S. Kim, S. B. Kwon, E. S. Park, C. H. Huh, S. W. Youn, S. W. Kim and K. C. Park, Protec-tive eff ects of EGCG on UVB-induced damage in living skin equivalents, Arch. Pharm. Res. 28 (2005) 784–790; DOI: 10.1007/BF02977343.
- M. Berneburg, H. Plett enberg, and J. Krutmann, Photoaging of human skin, Photodermatol. Photo-immunol. Photomed. 16 (2000) 239–244; DOI: 10.1034/j.1600-0781.2000.160601.x.